Difference between revisions of "QMCChem"
| Line 13: | Line 13: | ||
QMC=Chem is very well suited to massive parallelism and cloud computing: | QMC=Chem is very well suited to massive parallelism and cloud computing: | ||
| − | + | * All the implemented algorithms are CPU-bound | |
| − | + | * All workers are totally independent | |
| − | + | * The load balancing is optimal: the workers always work 100% of the time, independently of their respective CPU speeds | |
| − | + | * The code was written to be as portable as possible: the manager is written in standard Python and the worker is written in standard Fortran90 with MPI. | |
| − | + | * The network traffic is minimal and the amount of data transferred over the network can even be adjusted by the user | |
| − | + | * The number of simultaneous worker nodes can be variable during a calculation | |
| − | + | * Fault-tolerance can be easily implemented | |
| − | + | * The input and output data are not presented as traditional input files and output files. All the input and output data are stored in a database and an API is provided to access the data. This allows different forms of interaction of the user: scripts, graphical user interfaces, command-line tools, web interfaces, etc. | |
== Current Features == | == Current Features == | ||
Revision as of 18:06, 21 February 2011
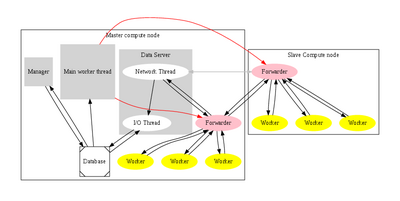
During the last three years we have been actively developing the QMC=Chem quantum Monte Carlo code. This code was initially designed for massively parallel simulations, and uses the manager/worker model.
When the program starts its execution, the manager runs on the master node and forks two other processes: a worker process and a data server. The worker is an efficient Fortran/MPI executable with minimal memory and disk space requirements (typically a few megabytes for each), where the only MPI communication is the broadcast of the input data (wave function parameters, initial positions in the 3N-space and random seed). The outline of the task of a worker is the following:
while ( Running )
{
compute_a_block_of_data();
Running = send_the_results_to_the_data_server();
}
The data server is an XML-RPC server implemented in Python. When it receives the computed data of a worker, it replies to the worker the order given by the manager to compute another block or to stop. The received data is then stored in a database using an asynchronous I/O mechanism. The manager is always aware of the results computed by all the workers and controls the running/stopping state of the workers and the interaction of the user during the simulation.
QMC=Chem is very well suited to massive parallelism and cloud computing:
- All the implemented algorithms are CPU-bound
- All workers are totally independent
- The load balancing is optimal: the workers always work 100% of the time, independently of their respective CPU speeds
- The code was written to be as portable as possible: the manager is written in standard Python and the worker is written in standard Fortran90 with MPI.
- The network traffic is minimal and the amount of data transferred over the network can even be adjusted by the user
- The number of simultaneous worker nodes can be variable during a calculation
- Fault-tolerance can be easily implemented
- The input and output data are not presented as traditional input files and output files. All the input and output data are stored in a database and an API is provided to access the data. This allows different forms of interaction of the user: scripts, graphical user interfaces, command-line tools, web interfaces, etc.
Contents
Current Features
Methods
- VMC
- DMC
- Jastrow factor optimization
- CI coefficients optimization
Wave functions
- Single determinant
- Multi-determinant
- Multi-Jastrow
- Nuclear cusp correction
Properties
Links
- Easy development with the IRPF90 tool.
- The access to input/output files is provided via an API produced by the Easy Fortran I/O library generator
- Documentation
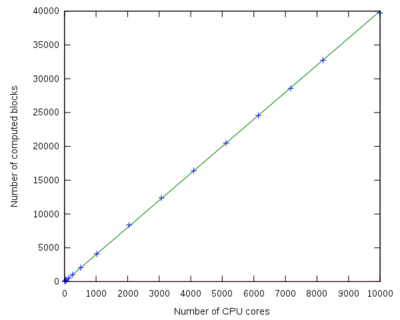
Parallel speed-up curve
| Number of processors | Number of computed blocks | Speed-up |
|---|---|---|
| 1 | 5 | 1 |
| 8 | 41 | 8 |
| 16 | 81 | 16 |
| 32 | 170 | 32 |
| 64 | 321 | 64 |
| 128 | 644 | 128 |
| 256 | 1280 | 256 |
| 512 | 2417 | 483 |
Features under development
Properties
- Molecular Forces
- Moments (dipole, quadrupole,...)
- Electron density
- ZV-ZB EPLF estimator
Practical aspects
- Web interface for input and output
Input file creation
The QMC=Chem input file can be created using the web interface. Upload a Q5Cost file or an output file from GAMESS, Gaussian or Molpro, and you will download the QMC=Chem input directory.